Bacterial biodiversity drives the evolution of CRISPR-based phage resistance
The paper I'll discuss in this brief post is this one: Bacterial biodiversity drives the evolution of CRISPR-based phage resistance (Ellinor O. Alseth, Elizabeth Pursey, Adela M. Luján, Isobel McLeod, Clare Rollie & Edze R. Westra, Nature 574, 549–552 (2019))
(The authors appear to be 100% women, nice to see.)
CRISPR is very much in the scientific news these days, both as a research tool and as a possible therapeutic agent for a host of genetic diseases, including (but hardly limited to) cancer, which may be thought of as a somatic genetic disease. With respect to its use as a research tool, a few days back I posted in this space, a report utilizing CRISPR to interrogate the toxin resistance that is observed in the Monarch butterfly. Genome editing retraces the evolution of toxin resistance in the monarch butterfly.
I haven't really paid much attention to the nuts and bolts of CRISPR technology, at least until a chance conversation at a science oriented social event stimulated me to do so. I was of course, aware of its role in gene therapy, but not of the basic science underlying it. The appearance of two papers in two subsequent issues of Nature stimulated more interest in the origins and use of this technology.
As many people know, among the many things we are leaving for future generations besides an atmosphere destroyed by appeals to denial, mysticism, fear and ignorance, and effective depletion of many of the elements in the periodic table, is a plethora of dangerous antibiotic resistant bacteria. One avenue for addressing this resistant bacteria is to appeal to an old idea, viral antibiotics, inoculating people with viruses that are known to attack and kill bacteria, viruses know as phages. (Phages are also widely used as research and production tools, particularly for the insertion of genes into organisms in the biotech industry.) This old idea is worth a look given that our understanding of molecular biology has entered a golden age which, one hopes, will be maintained despite the rise of anti-intellectualism on both political extremes.
Anyway.
The CRISPR/CAS-9 system is actually the immune system for bacteria, and this paper is about how this system is utilized to develop resistance to phages.
From the abstract of the paper:
About half of all bacteria carry genes for CRISPR–Cas adaptive immune systems1, which provide immunological memory by inserting short DNA sequences from phage and other parasitic DNA elements into CRISPR loci on the host genome2. Whereas CRISPR loci evolve rapidly in natural environments3,4, bacterial species typically evolve phage resistance by the mutation or loss of phage receptors under laboratory conditions5,6. Here we report how this discrepancy may in part be explained by differences in the biotic complexity of in vitro and natural environments7,8. Specifically, by using the opportunistic pathogen Pseudomonas aeruginosa and its phage DMS3vir, we show that coexistence with other human pathogens amplifies the fitness trade-offs associated with the mutation of phage receptors, and therefore tips the balance in favour of the evolution of CRISPR-based resistance. We also demonstrate that this has important knock-on effects for the virulence of P. aeruginosa, which became attenuated only if the bacteria evolved surface-based resistance. Our data reveal that the biotic complexity of microbial communities in natural environments is an important driver of the evolution of CRISPR–Cas adaptive immunity, with key implications for bacterial fitness and virulence.
From the introduction:
P. aeruginosa is a widespread opportunistic pathogen that thrives in a range of different environments, including hospitals, where it is a common source of nosocomial infections. In particular, it frequently colonizes the lungs of patients with cystic fibrosis, in whom it is the leading cause of morbidity and mortality9. In part fuelled by a renewed interest in the therapeutic use of bacteriophages as antimicrobials (phage therapy)10,11, many studies have examined whether and how P. aeruginosa evolves resistance to phage (reviewed in ref. 12). The clinical isolate P. aeruginosa strain PA14 has been reported to predominantly evolve resistance against its phage DMS3vir by the modification or complete loss of the phage receptor (type IV pilus) when grown in nutrient-rich medium5, despite carrying an active CRISPR–Cas adaptive immune system. By contrast, under nutrient-limited conditions, the same strain relies on CRISPR–Cas to acquire phage resistance5. These differences are due to higher phage densities during infections in nutrient-rich compared with nutrient-limited conditions, which in turn determines whether surface-based resistance (with a fixed cost of resistance) or CRISPR-based resistance (infection-induced cost) is favoured by natural selection5,13. Although these observations suggest abiotic factors are crucial determinants of the evolution of phage resistance strategies, the role of biotic factors has remained unclear, even though P. aeruginosa commonly co-exists with a range of other bacterial species in both natural and clinical settings14,15. We proposed that the presence of a bacterial community could drive increased levels of CRISPR-based resistance evolution for two main reasons. First, reduced densities of P. aeruginosa in the presence of competitors may limit phage amplification, and favour CRISPR-based resistance5. Second, pleiotropic costs associated with the mutation of phage receptors may be amplified during interspecific competition.
The authors used in their experiments, a streptomycin resistant strain (PA14) of the
P. aeruginosa pathogenic organism and cutlured it in the presence of three other pathogenic bacterial species that are not infected by the DMS3vir virus,
Staphylococcus aureus, Burkholderia cenocepacia and
Acinetobacter baumannii.
Some pictures from the paper suggesting the results:
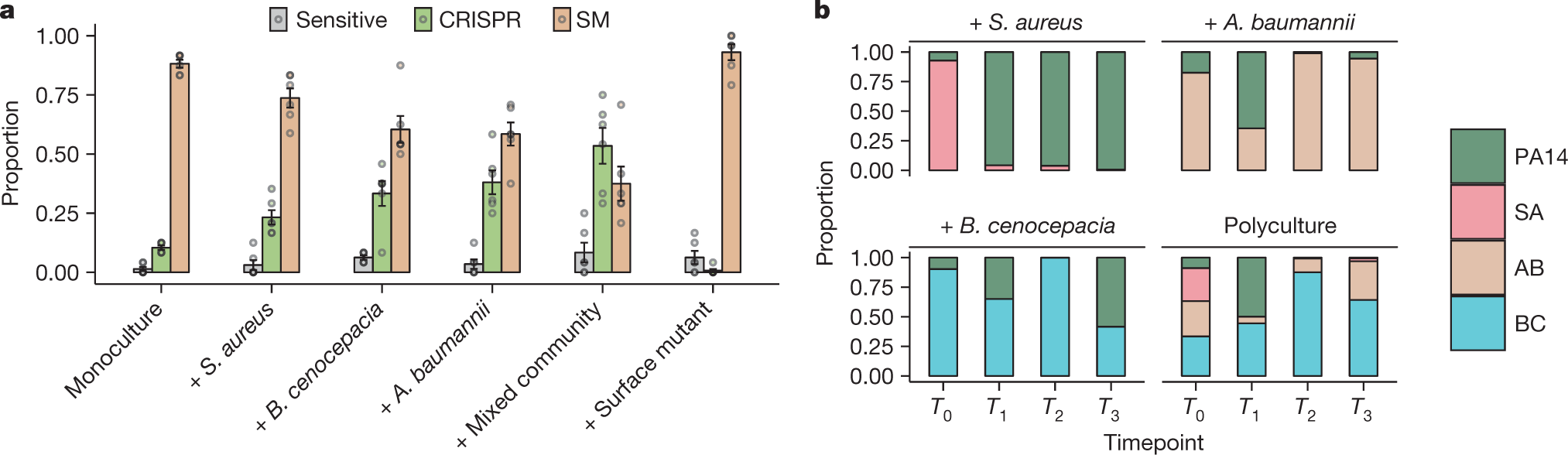
The caption:
a, Proportion of P. aeruginosa that acquired surface modification (SM) or CRISPR-based resistance, or remained sensitive at 3 d.p.i. with phage DMS3vir when grown in monoculture or polycultures, or with an isogenic surface mutant (6 replicates per treatment, with 24 colonies per replicate, n = 36 biologically independent replicates). Data are mean ± s.e.m. b, Microbial community composition over time for the mixed-species infection experiments. AB, A. baumannii; BC, B. cenocepacia; PA14, P. aeruginosa; SA, S. aureus.
The authors also evaluated the potential pathogenic implications of this result by growing cultures in synthetic sputum.
The rise of CRISPR resistance, as shown in the previous graphic in
P. aeruginosa in the presence of bacterial diversity is clearly observed.
Another graphic:
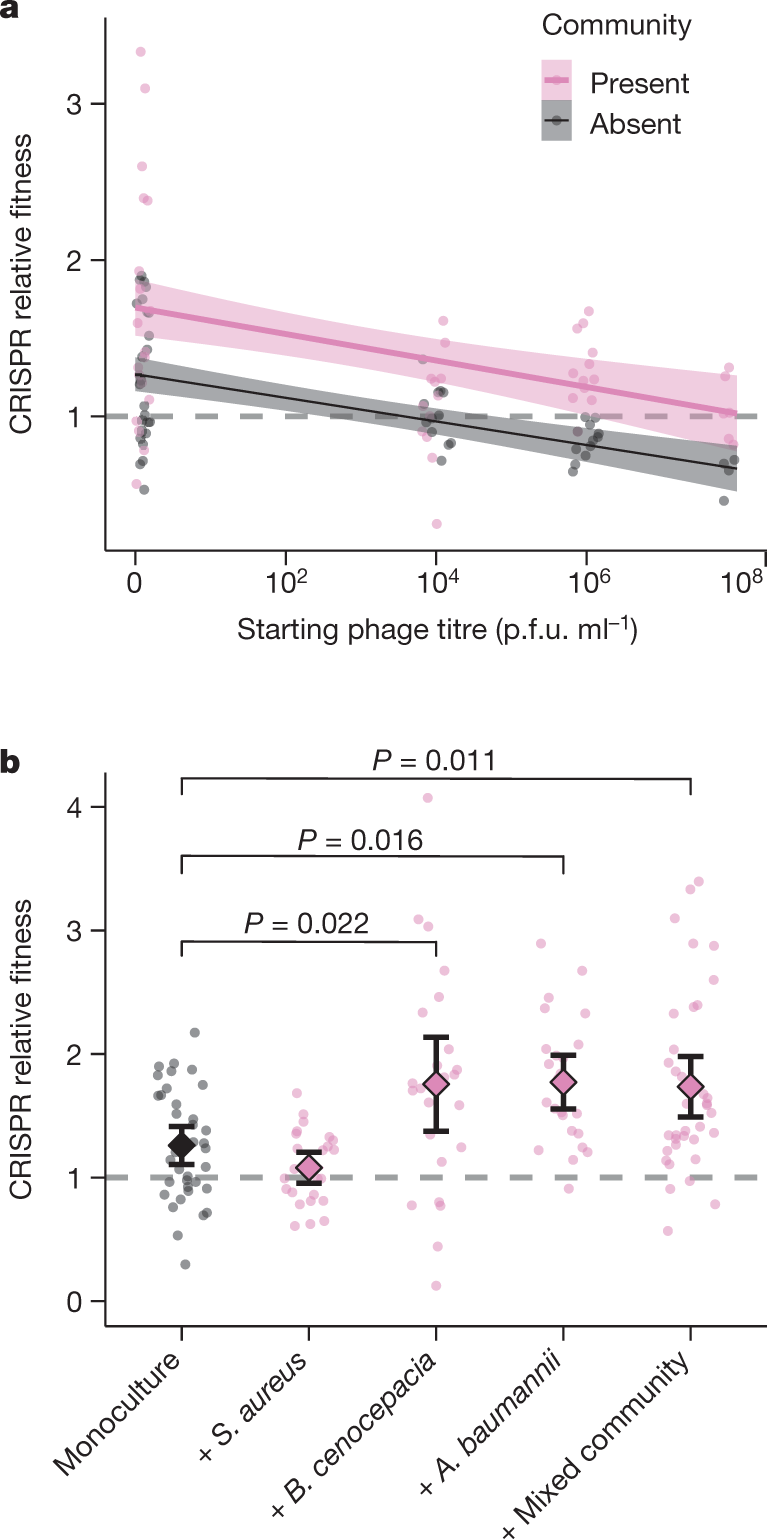
The caption:
a, Relative fitness of a P. aeruginosa clone with CRISPR-based resistance after competing for 24 h against a surface-modification clone at varying titres of phage DMS3vir in the presence or absence of a mixed microbial community. Regression slopes with shaded areas corresponding to 95% confidence interval (n = 144 biologically independent samples). b, Relative fitness after competition in the absence of phage, but in the presence of other bacterial species individually or as a mixture. Data are mean and 95% confidence intervals (n = 144 biologically independent samples).
Another graphic touching on virulence:
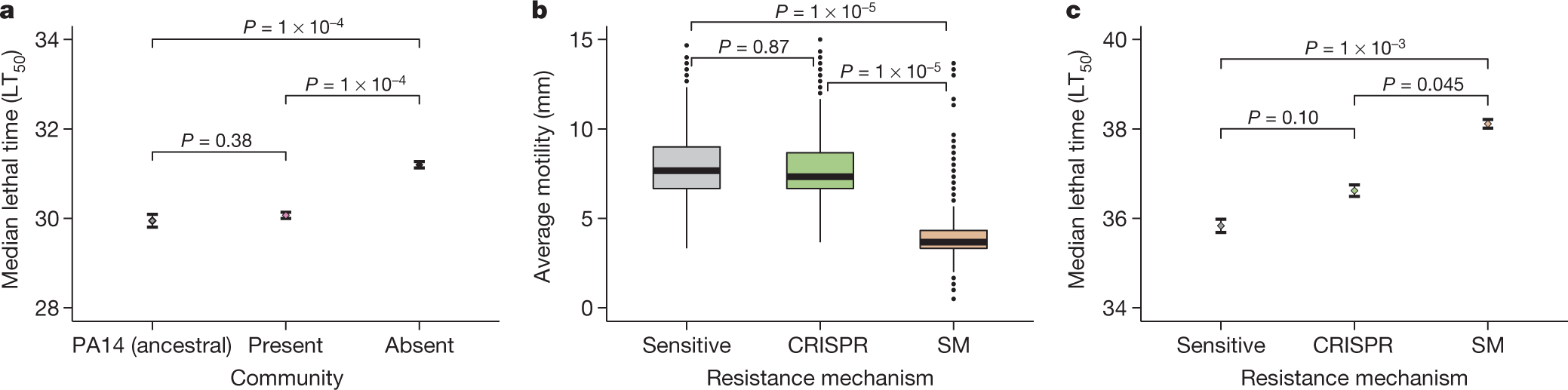
a, Time until death (given as the median ± one standard error) after infection with PA14 clones that evolved phage resistance in either the presence or the absence of a mixed microbial community (n = 376 biologically independent samples, analysed using a Cox proportional-hazards model with Tukey contrasts). LT50, median lethal time. b, The effect of the type of evolved phage resistance (CRISPR-based or surface-modification-based) on bacterial motility (n = 981 biologically independent samples). Box plots show the median with the upper and lower twenty-fifth and seventy-fifth percentiles, the interquartile range, and outliers shown as dots. c, The effect of the type of resistance on in vivo virulence (time until death, given as the median ± one standard error; n = 981, analysed using a Cox proportional-hazards model with Tukey contrasts).
Some excerpts from the concluding discussion:
We have shown that the evolutionary outcome of bacteria–phage interactions can be fundamentally altered by the microbial community context. Although conventionally studied in isolation, these interactions are usually embedded in complex biotic networks of interacting species, and it is becoming increasingly clear that this can have key implications for the evolutionary epidemiology of infectious disease24,25,26,27,28. Our work shows that the community context can shape the evolution of different host-resistance strategies. Specifically, we find that the interspecific interactions between four bacterial species in a synthetic microbial community can have a large effect on the evolution of phage-resistance mechanisms by amplifying the constitutive fitness cost of surface-based resistance5. The finding that biotic complexity matters complements previous work on the effect of abiotic variables and force of infection on the evolution of phage resistance5...
...Primarily, the absence of detectable trade-offs between CRISPR-based resistance and virulence, as opposed to when bacteria evolve surface-based resistance, suggests that the evolution of CRISPR-based resistance can ultimately influence the severity of disease. Moreover, the evolution of CRISPR-based resistance can drive more rapid phage extinction29, and may in a multi-phage environment result in altered patterns of cross-resistance evolution compared with surface-based resistance30. The identification of the drivers and consequences of CRISPR-resistance evolution might help to improve our ability to predict and manipulate the outcome of bacteria–phage interactions in both natural and clinical settings.
Interesting, I think, and important.
Have a nice day tomorrow.


